
The research of our laboratory focuses on lead discovery and optimization using synthetic chemistry in close conjunction with liquid chromatography coupled to mass spectrometry (LC-MS) and molecular modeling. We are interested in the development of straightforward strategies for fragment-based lead discovery and optimization using the protein of interest to assemble its own bidentate ligand from a library of complementary reacting fragments. In addition, we are developing hit-to-lead progression strategies conducting structure-activity relationship (SAR) and structure-property relationship (SPR) studies in parallel. We are currently applying our lead discovery approaches for the discovery and optimization of therapeutic agents targeting cancer and infectious diseases. The second research pillar of our laboratory comprises the development of chemical tools to label and identify specific proteins in complex mixtures or entire proteomes. We are focusing on the development of various probes to investigate proteins related to cell-cell communication and energy metabolism.
Kinetic Target-Guided Synthesis: In the last decade, fragment-based lead discovery or kinetic Target-Guided Synthesis (TGS) approaches have been developed in which the biological target is actively engaged in the design and the synthesis of its own inhibitory compounds. In these fragment-based lead discovery methods, the biological target – an enzyme or a DNA fragment – is directly involved in the irreversible assembly of its own bidentate ligand from a pool of smaller reactive fragments (Figure 1). The Manetsch laboratory is interested in the development of kinetic TGS approaches and in the study of their scope as a lead discovery and/or optimization tool. Our ideas are aimed at developing synthesis and screening methods that generate only biologically active compounds. Currently, the Manetsch laboratory is applying kinetic TGS targeting various enzymatic targets and protein-protein interactions.
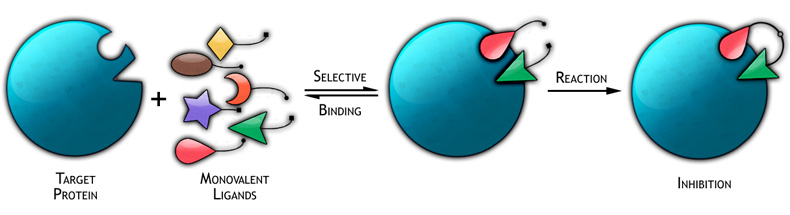 Figure 1 |
Protein-protein interactions are central to many biological processes and hence represent a large and important class of targets for human therapeutics. Recent discovery of a variety of low-molecular-weight compounds that interfere with biologically relevant protein-protein complexes has fostered the identification and validation of new therapy strategies for a variety of diseases. Nevertheless, disrupting or modulating protein-protein interactions with low-molecular-weight compounds remains challenging due to the lack of deep binding pockets on protein surfaces. Moreover, the adaptive nature of binding sites on protein surfaces creates additional challenges for lead compound design and drug discovery. Despite successful discovery of several Protein-Protein Interaction Modulators (PPIMs) in recent years, there is an urgent need to find simple, yet highly reliable and less cumbersome approaches to PPIM discovery. In a proof-of-concept study using the protein Bcl-xL, a central regulator of programmed cell death, we demonstrated that an amidation reaction between thio acids and sulfonyl azides is applicable for Bcl-xL-templated assembly of inhibitory compounds (Figure 2). We have shown for the first time that kinetic TGS can be applied not only on enzymatic targets but also for the discovery of small molecules modulating protein−protein interactions. (Article Link)
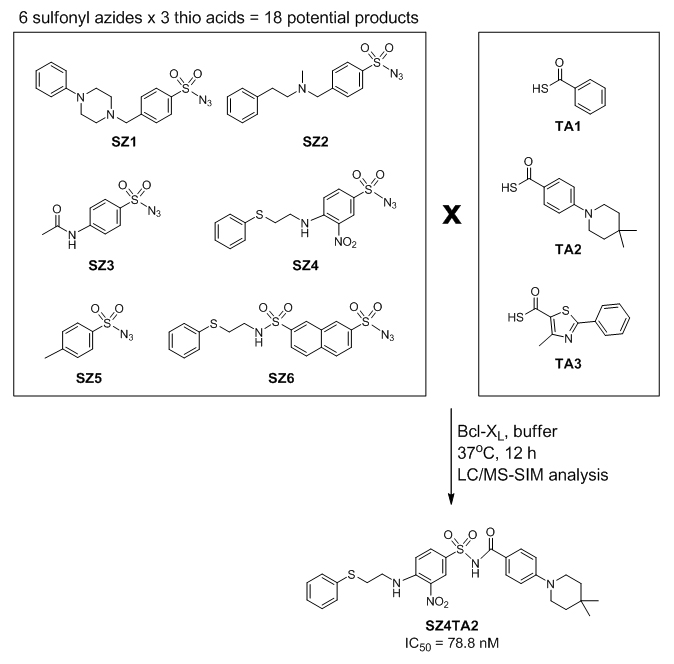 Figure 2 |
Hit-to-Lead Progression Strategies: Another research activity of the Manetsch laboratory is lead design and optimization targeting infectious diseases such as influenza, malaria and Leishmania. Analogues of hit compounds identified through High-Throughput Screening (HTS) are synthesized and tested with the intention to improve and optimize the inhibitory enabling to obtain solid Structure-Activity Relationship (SAR) data. In addition, basic physical drug properties such as solubility, permeability, logD and metabolic stability are routinely determined in order to obtain Structure-Property Relationship (SPR) data of each individual compound tested for inhibitory activity. SAR and SPR data is carefully taken into consideration in the iterative process of inhibitor design and optimization. Various LC/MS-based SPR assays have been implemented in the Manetsch laboratory.
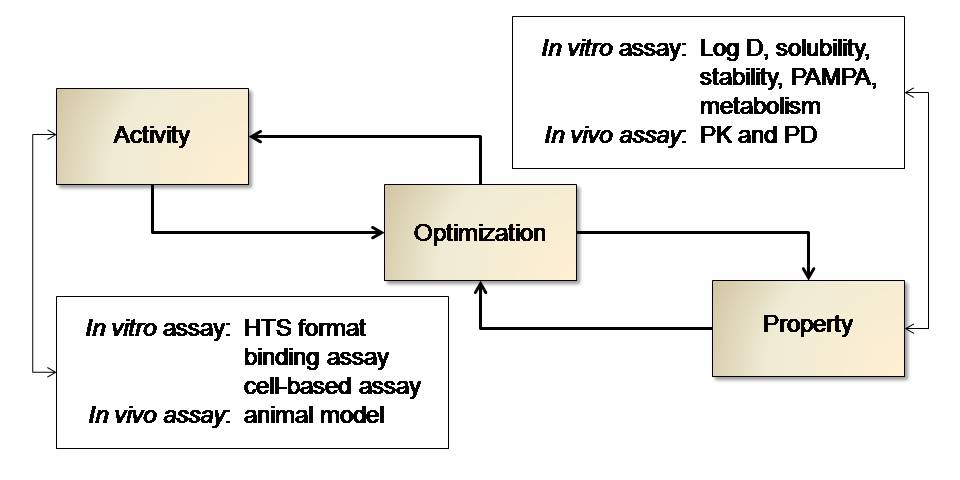 Figure 3 |
4(1H)-quinolones such as endochin (1) and ICI56,780 (2) are known causal prophylactic (kill growing liver stage parasites) and potent erythrocytic stage agents in avian malaria models, but not against malaria parasites in mammals (Figure 4). These observations stimulated us to evaluate quinolone derivatives obtained from the Walter Reed Army Institute of Research chemical inventory in current in vitro screening platform against the atovaquone (3) resistant P. falciparum parasites W2 and TM90-C2B. Surprisingly, several 1,2,3,4-tetrahydroacridones and 3-substituted 2-methyl-4(1H)-quinolones displayed not only remarkable erythrocytic stage activity, but the cross resistance with atovaquone (3) was not found to be complete across the chemical series. Using optimized palladium-catalyzed cross coupling reaction conditions, we devised synthetic strategies enabling the rapid and reliable preparation of libraries of differently substituted 4(1H)-quinolones (4) and 1,2,3,4-tetrahydroacridones (5). From all the synthesized and tested compounds, we have discovered quinolones and tetrahydroacridones preventing growth of P. falciparum at single digit nM concentrations in vitro. Currently, DMPK studies as well as mechanism of action studies of these novel chemotypes are ongoing.
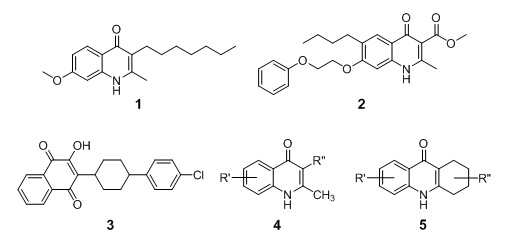 Figure 4 |
Protein Labeling Tools: Adenine nucleosides and nucleotides (ADoR's) are key molecules in a large number of metabolic and cell signaling pathways. Thus, ADoR-dependent proteins correlate many human diseases including cancer, cardiovascular diseases, diabetes, and obesity. The Manetsch laboratory developed reliable synthetic strategies for the modular preparation of biotinylated azido-adenosine analogues for the study of ADoR's-dependent proteomes. The design of these AdoR probes includes a photolabile azide moiety for covalent attachment of the probe to the target proteins as well as a biotin moiety for the isolation of the probe-protein conjugates using avidin affinity chromatography (Figure 5). We are currently investigation the prowess of our photoaffinity probes on different proteomes.
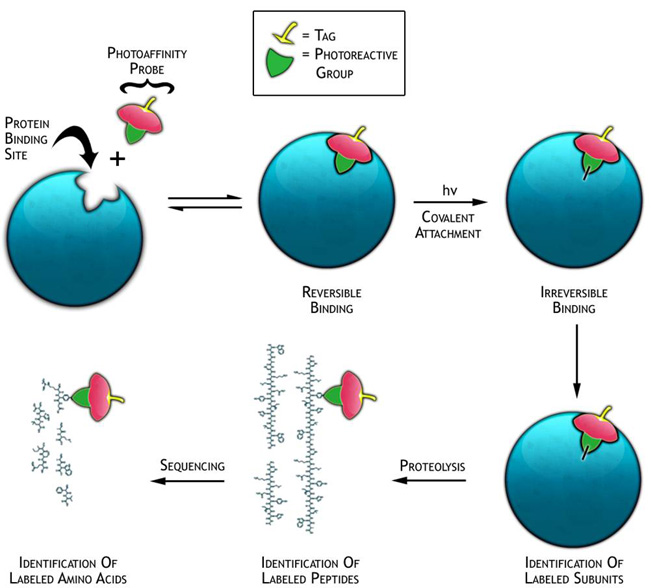 Figure 5 |
|
Collaborators of Manetsch Laboratory: John H. Adams Department of Global Health College of Public Health University of South Florida 3720 Spectrum Blvd, IDRB 304/office 331 Tampa, FL 33612 Office: (813) 974-9916 Lab: (813) 974-0799 or 4-0656 jadams3 [at] health.usf.edu Website Susan A. Charman Centre for Drug Candidate Optimisation Monash Institute of Pharmaceutical Sciences Monash University (Parkville Campus) 381 Royal Parade Parkville, Victoria 3052 Australia Phone: +61 3 9903 9626 Susan.Charman [at] pharm.monash.edu.au Website Gloria Ferreira Department of Molecular Medicine College of Medicine University of South Florida 12901 Bruce B. Downs Blvd., MDC 2152 Tampa, Florida 33612 Phone: (813) 974-2332 aseyfang [at] health.usf.edu Website Dennis E. Kyle Department of Global Health College of Public Health University of South Florida 3720 Spectrum Blvd, IDRB 304 Tampa, FL 33612 Office: (813) 974-1273 Lab: (813) 974-7191 or 4-4578 dkyle [at] health.usf.edu Website David J. Merkler Department of Chemistry University of South Florida 4202 E. Fowler Avenue, BSF 307 Tampa, FL 33620-5250 Office: (813) 974-3579 Email: merkler [at] cas.usf.edu Website Andreas Seyfang Department of Molecular Medicine College of Medicine University of South Florida 12901 Bruce B. Downs Blvd., MDC Box 7 Tampa, Florida 33612 Phone: (813) 974-2332 aseyfang [at] health.usf.edu Website Alberto L. van Olphen USF-Center for Biological Defense (CBD) Global Health Infectious Disease Research (GHIDR) Department of Global Health MDC56 College of Public Health University of South Florida 3720 Spectrum Boulevard, IDRB 304 Tampa, FL 33612 Phone: (813) 974-8794 avanolph [at] health.usf.edu Hong-Gang Wang Penn State College of Medicine Department of Pharmacology, R130 500 University Drive, P. O. Box 850 Hershey, PA 17033-0850 Phone: 717-531-0003 (X 285881) hwang3 [at] hmc.psu.edu |
|
Funding Agencies: Medicines for Malaria Venture James and Esther King Foundation  Bankhead-Coley  Johhnie B. Byrd, Sr. Alzheimer's Center and Research Institute 
|
